Recently I came across a machine learning algorithm called 'k-nearest neighbors' or 'kNN,' which is used as a predictive modeling tool. This algorithm uses data to build a model and then uses that model to predict the outcome.
kNN is new for me, and I gained most of my knowledge by reading these two tutorials; tutorial 1 and tutorial 2.
I will apply the kNN algorithm in the NHANES data set to predict diabetes.
Load 'tidyverse,' 'class,' and 'NHANES' packages.
library(tidyverse)
library(RNHANES)
library(class)
Import the dataset
DEMO_F = nhanes_load_data("DEMO_F", "2009-2010") %>%
select(SEQN, RIDAGEYR)
BMX_F = nhanes_load_data("BMX_F", "2009-2010") %>%
select(SEQN, BMXBMI, BMXWT)
HDL_F = nhanes_load_data("HDL_F", "2009-2010") %>%
select(SEQN, LBDHDD)
GLU_F = nhanes_load_data("GLU_F", "2009-2010") %>%
select(SEQN, LBXGLU, LBXIN)
DIQ_F = nhanes_load_data("DIQ_F", "2009-2010") %>%
select(SEQN, DIQ010)
Merge the data in one dataset and remove missing values.
dtx = left_join(DEMO_F, HDL_F) %>%
left_join(GLU_F) %>%
left_join(BMX_F) %>%
left_join(DIQ_F)
dat = dtx %>%
filter(!is.na(BMXBMI), !is.na(LBDHDD), !is.na(LBXGLU), !is.na(LBXIN),RIDAGEYR >= 40, DIQ010 %in% c(1, 2)) %>%
transmute(SEQN, Age = RIDAGEYR, BMI = BMXBMI, Cholest = LBDHDD, Glucose = LBXGLU, Insuline = LBXIN, Weight = BMXWT, Diabetes = DIQ010) %>%
mutate(Diabetes = recode_factor(Diabetes,
`1` = "Yes",
`2` = "No"))
Explore data
Now that I have the dataset, I will evaluate the correlation between two variables (glucose and weight) using the 'ggplot2' package.
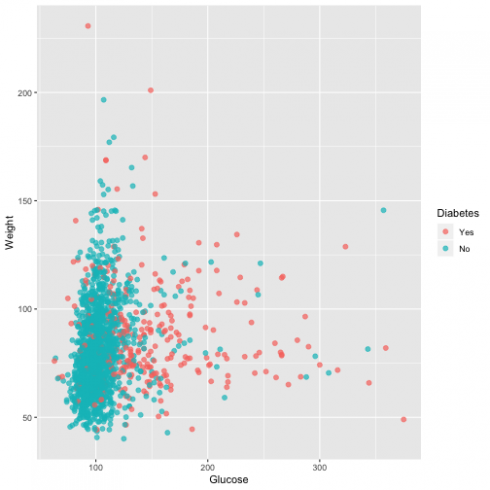
ggplot(dat, aes(Glucose, Weight, color = Diabetes)) +
geom_point(alpha = 0.7, size = 2)
From the graph, it is evident that the higher the level of blood glucose more diabetes events we have.
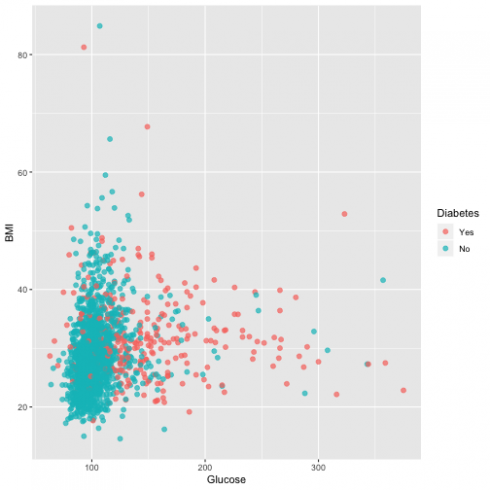
Another relationship I want to assess is Blood glucose and BMI:
ggplot(dat, aes(Glucose, BMI, color = Diabetes)) +
geom_point(alpha = 0.7, size = 2)
Prepare the data set
To proceed with preparing the data, I would like to get more information regarding all variables, and I use 'summary' function:
summary(dat)
## SEQN Age BMI Cholest Glucose
## Min. :51645 Min. :40.00 Min. :14.59 Min. : 19.00 Min. : 63
## 1st Qu.:54366 1st Qu.:48.00 1st Qu.:24.96 1st Qu.: 43.00 1st Qu.: 95
## Median :57096 Median :59.00 Median :28.31 Median : 52.00 Median :102
## Mean :56983 Mean :59.23 Mean :29.36 Mean : 54.66 Mean :111
## 3rd Qu.:59593 3rd Qu.:70.00 3rd Qu.:32.62 3rd Qu.: 64.00 3rd Qu.:114
## Max. :62158 Max. :80.00 Max. :84.87 Max. :144.00 Max. :375
## Insuline Weight Diabetes
## Min. : 0.880 Min. : 40.10 Yes: 286
## 1st Qu.: 7.008 1st Qu.: 66.90 No :1486
## Median : 11.405 Median : 78.15
## Mean : 15.101 Mean : 81.73
## 3rd Qu.: 17.995 3rd Qu.: 92.92
## Max. :320.220 Max. :230.70
Here I see a different range of values in my variables; therefore, I will normalize my numeric variables (the predictors) to prepare them for using in the kNN algorithm. I create the normalize function and then apply it to the predictor variables as below:
normalize <- function (i) {
(i - min(i))/(max(i) - min(i))
}
norm_dat <- dat %>%
select(Age, BMI, Cholest, Glucose, Insuline, Weight) %>%
lapply(., normalize) %>%
as.data.frame()
All the new values are within the range of 0 and 1.
summary(norm_dat)
## Age BMI Cholest Glucose Insuline
## Min. :0.0000 Min. :0.0000 Min. :0.0000 Min. :0.0000 Min. :0.00000
## 1st Qu.:0.2000 1st Qu.:0.1476 1st Qu.:0.1920 1st Qu.:0.1026 1st Qu.:0.01919
## Median :0.4750 Median :0.1952 Median :0.2640 Median :0.1250 Median :0.03296
## Mean :0.4807 Mean :0.2102 Mean :0.2853 Mean :0.1539 Mean :0.04453
## 3rd Qu.:0.7500 3rd Qu.:0.2565 3rd Qu.:0.3600 3rd Qu.:0.1635 3rd Qu.:0.05359
## Max. :1.0000 Max. :1.0000 Max. :1.0000 Max. :1.0000 Max. :1.00000
## Weight
## Min. :0.0000
## 1st Qu.:0.1406
## Median :0.1996
## Mean :0.2184
## 3rd Qu.:0.2772
## Max. :1.0000
To evaluate my kNN model's performance, I will divide the data set into a training and a test set. In the training dataset, I will perform the algorithm, and the test set will serve to asses the algorithm. The split must be 2/3 for a train set and 1/3 for the test set, and I need to make sure that both data sets have the same ratio of 'having or not' diabetes participants.
I use the 'sample()' function to take a sample with the same size that is my dataset 'dat' and assign 1 or 2 to each row with probability 0.67 and 0.33. Then, use it to define my training and test data sets:
dat_samp <- sample(2, nrow(dat), replace=TRUE, prob=c(0.67, 0.33))
dat_training <- norm_dat[dat_samp==1, 1:6]
dat_test <- norm_dat[dat_samp==2, 1:6]
I will store the 8-th column of my train data, which is the target variable (diabetes) in 'dat_target_group' because it will be used as 'cl' argument in knn function. Also, store the 8-th column of my test set in 'dat_target_group,' which I will use later to test the accuracy of the algorithm used.
dat_target_group <- dat[dat_samp==1, 8]
dat_test_group <- dat[dat_samp==2, 8]
Run the kNN algorithm
dat_pred <- knn(train = dat_training, test = dat_test, cl = dat_target_group, k=3)
dat_pred
## [1] No No No No No Yes No No No No Yes No No Yes No No No No No No No
## [22] Yes No No No No Yes No No No No Yes No No No No No No No No No No
## [43] No No Yes No Yes No No No No No No No Yes No No Yes No No No No No
## [64] No No No No No No No Yes No No No Yes No No No No No No Yes No No
## [85] No No No No Yes No No No No No No No No No No No No No No Yes No
## [106] Yes No No No No No No No No Yes No No No No No No No No No No Yes
## [127] No No No No No No No No No No No No No No No No No No No No No
## [148] No No No No Yes No No Yes No No No No Yes No No No Yes No No No No
## [169] No No No No No No No No Yes No No No Yes No No Yes No No No No No
## [190] No Yes No No No No No Yes No No No No No No No No No No Yes No No
## [211] No No No No No No No No No No No No No No No No No No No No No
## [232] No No No No No No No No No No Yes No No No No No No No Yes No No
## [253] Yes No No No No Yes No No No No No No No No No Yes No No No No No
## [274] No Yes Yes No Yes No No No No No No No No No Yes No No No No No No
## [295] No No No No No No No No No No No No No No No No Yes No No No No
## [316] No No No No No No No No No No No No No No No No No No No No No
## [337] No No No No No No No No No No No No No No No Yes No No No No No
## [358] Yes No No No No Yes No No No No No No No Yes Yes No No No No No No
## [379] No No No No No Yes No No No No No No No No No No No Yes No No No
## [400] No No No No No No No No No No No No Yes No No No No No No No No
## [421] No No No No No No No No No No No No No Yes No No No No No No No
## [442] No No No No No No No No No No No No No No No No No No No No No
## [463] No No No No No No No No No No No Yes Yes Yes No No No No No Yes No
## [484] No No No No Yes No No No No No No No No No No No No No No No No
## [505] No No No No Yes No No No No No No No No No Yes Yes Yes No No Yes Yes
## [526] No No No No No No No No No No
## Levels: Yes No
These are my predictive values. Note that the k parameter is often an odd number and means the amount of nearest neighbors you decide to check for every participant to determine in what category (Yes/No) for diabetes will he be assigned.
Below I can check the participants with and without diabetes in both datasets:
summary(dat_pred)
## Yes No
## 58 477
summary(dat_test_group)
## Yes No
## 86 449
Evaluate the model
To evaluate my kNN model I build a specific table (confusion matrix) with 'table' function putting my predictive values 'dat_pred' and values from my test set 'dat_test_group' to asses the kNN model.
tab <- table(dat_pred, dat_test_group)
tab
## dat_test_group
## dat_pred Yes No
## Yes 37 21
## No 49 428
Now I create the 'accuracy' function and use it to evaluate 'tab' :
accuracy <- function(x){sum(diag(x)/(sum(rowSums(x)))) * 100}
accuracy(tab)
## [1] 86.91589
In the NHANES dataset, I have run the k-nearest neighbor algorithm that gave me an 87% accurate.
As I mentioned in the top of the post, I am new to kNN so please suggest or correct my code.