In this post, I am going to build a statistical learning model as based upon plant leaf datasets introduced in part one of this tutorial. We have available three datasets, each one providing sixteen samples each of one-hundred plant species.
The features are:
- shape
- texture
- margin
Specifically, I will take advantage of Discrimination Analysis for classification purposes and I will compare my results with the ones of ref. [1] table 2, as herein reported.
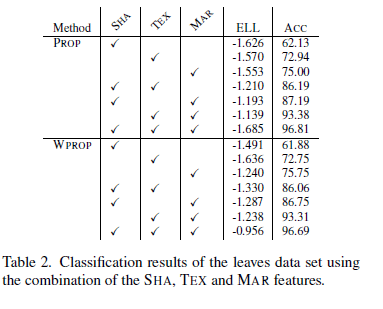
Ref. [1] authors show the accuracy reached by KNN (proportional and weighted proportional kernel density) for any combination of the datasets in use. Their top accuracy is 96% and it is achieved when using all three datasets. In the present tutorial I am going to compare my results with the ones obtained in ref. [1].
Packages
suppressPackageStartupMessages(library(caret))
suppressPackageStartupMessages(library(dplyr))
suppressPackageStartupMessages(library(ggplot2))
CopyClassification Models
Loading previous post environment where datasets are available.
load('PlantLeafEnvironment.RData')
CopyWe define an utility function to gather all the steps needed for model building by caret package (ref. [3]), specifically:
- train partition as 70% training and 30% test samples
- train control specifying repeated cross-validation with ten folds
- features set, all features besides species and id
- train dataset
- test dataset
- linear discriminant analysis fit, with preprocessing to compensate for spatial distribution asymmetries and to center and scale values
- prediction computation
- confusion matrix computed on the test dataset
- result as made of the linear discriminant analysis fit and the test dataset confusion matrix
As pre-processing step, also PCA (Principal Component Analysis) is specified. That allows for slightly better accuracy results compared to non PCA preprocessing scenario (see also ref. [4] for further insights about combining PCA and LDA).
Among all available Discriminant Analysis models within caret package, the lda appears to be the most efficient with time and as well providing with good results, as we will see at the end. For details about Linear Discriminant Analysis, please see ref. [5] and [6].
set.seed(1023)
plant_leaf_model <- function(leaf_dataset) {
train_idx <- createDataPartition(leaf_dataset$species, p = 0.7, list = FALSE)
trControl <- trainControl(method = "repeatedcv", number = 10, verboseIter = FALSE, repeats = 5)
relevant_features <- setdiff(colnames(leaf_dataset), c("id", "species"))
feat_sum <- paste(relevant_features, collapse = "+")
frm <- as.formula(paste("species ~ ", feat_sum))
train_set <- leaf_dataset[train_idx,]
test_set <- leaf_dataset[-train_idx,]
lda_fit <- train(frm,
data = train_set,
method ="lda",
tuneLength = 10,
preProcess = c("BoxCox", "center", "scale", "pca"),
trControl = trControl,
metric = 'Accuracy')
lda_test_pred <- predict(lda_fit, test_set)
cfm <- confusionMatrix(lda_test_pred, test_set$species)
list(model=lda_fit, confusionMatrix = cfm)
}
CopyMargin dataset model
We build our model for margin dataset only.
result <- plant_leaf_model(margin_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 64 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: centered (64), scaled (64), principal component
## signal extraction (64)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1082, 1076, 1078, 1081, 1086, 1079, ...
## Resampling results:
##
## Accuracy Kappa
## 0.7947299 0.7924885
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.7950000 0.7929293 0.7520681 0.8335033
CopyWe take notice of accuracy for final report.
margin_data_accuracy <- result$confusionMatrix$overall[1]
CopyFurther details can be printed out by:
result$model$finalModel
varImp(result$model)
Copythat we do not show for brevity.
Shape dataset model
We build our model for shape dataset only.
result <- plant_leaf_model(shape_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 64 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: Box-Cox transformation (64), centered (64), scaled
## (64), principal component signal extraction (64)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1080, 1072, 1074, 1084, 1076, 1087, ...
## Resampling results:
##
## Accuracy Kappa
## 0.5167374 0.5116926
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.5150000 0.5101010 0.4648181 0.5649584
CopyWe take notice of accuracy for final report.
shape_data_accuracy <- result$confusionMatrix$overall[1]
CopyTexture dataset model
We build our model for texture dataset only.
result <- plant_leaf_model(texture_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 64 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: centered (64), scaled (64), principal component
## signal extraction (64)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1078, 1078, 1086, 1076, 1076, 1082, ...
## Resampling results:
##
## Accuracy Kappa
## 0.7580786 0.7554372
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.7575000 0.7550505 0.7124314 0.7987117
CopyWe take notice of accuracy for final report.
texture_data_accuracy <- result$confusionMatrix$overall[1]
CopyMargin+Shape datasets model
We build our model for margin and shape datasets.
margin_shape_data <- left_join(margin_data, shape_data[,-which(colnames(texture_data) %in% c("species"))], by=c("id"))
result <- plant_leaf_model(margin_shape_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 128 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: Box-Cox transformation (64), centered (128), scaled
## (128), principal component signal extraction (128)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1082, 1084, 1074, 1087, 1084, 1081, ...
## Resampling results:
##
## Accuracy Kappa
## 0.9511508 0.9506051
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.9300000 0.9292929 0.9004175 0.9529847
CopyWe take notice of accuracy for final report.
margin_shape_data_accuracy <- result$confusionMatrix$overall[1]
CopyMargin+Texture datasets model
We build our model for margin and texture datasets.
margin_texture_data <- left_join(margin_data, texture_data[,-which(colnames(texture_data) %in% c("species"))], by=c("id"))
result <- plant_leaf_model(margin_texture_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 128 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: centered (128), scaled (128), principal component
## signal extraction (128)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1079, 1078, 1084, 1074, 1085, 1081, ...
## Resampling results:
##
## Accuracy Kappa
## 0.9666299 0.9662565
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.9475000 0.9469697 0.9208654 0.9672116
CopyWe take notice of accuracy for final report.
margin_texture_data_accuracy <- result$confusionMatrix$overall[1]
CopyShape+Texture datasets model
We build our model for shape and texture datasets.
shape_texture_data <- left_join(shape_data, texture_data[,-which(colnames(texture_data) %in% c("species"))], by=c("id"))
result <- plant_leaf_model(shape_texture_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 128 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: Box-Cox transformation (64), centered (128), scaled
## (128), principal component signal extraction (128)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1083, 1080, 1080, 1082, 1082, 1081, ...
## Resampling results:
##
## Accuracy Kappa
## 0.9102814 0.9092814
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.9225000 0.9217172 0.8917971 0.9467373
CopyWe take notice of accuracy for final report.
shape_texture_data_accuracy <- result$confusionMatrix$overall[1]
CopyMargin+Shape+Texture datasets model
We build our model for all three datasets.
leaves_data <- left_join(margin_data, shape_texture_data[,-which(colnames(texture_data) %in% c("species"))], by = c("id"))
result <- plant_leaf_model(leaves_data)
CopyModel fit results.
result$model
## Linear Discriminant Analysis
##
## 1200 samples
## 192 predictor
## 100 classes: 'Acer Campestre', 'Acer Capillipes', 'Acer Circinatum', 'Acer Mono', 'Acer Opalus', 'Acer Palmatum', 'Acer Pictum', 'Acer Platanoids', 'Acer Rubrum', 'Acer Rufinerve', 'Acer Saccharinum', 'Alnus Cordata', 'Alnus Maximowiczii', 'Alnus Rubra', 'Alnus Sieboldiana', 'Alnus Viridis', 'Arundinaria Simonii', 'Betula Austrosinensis', 'Betula Pendula', 'Callicarpa Bodinieri', 'Castanea Sativa', 'Celtis Koraiensis', 'Cercis Siliquastrum', 'Cornus Chinensis', 'Cornus Controversa', 'Cornus Macrophylla', 'Cotinus Coggygria', 'Crataegus Monogyna', 'Cytisus Battandieri', 'Eucalyptus Glaucescens', 'Eucalyptus Neglecta', 'Eucalyptus Urnigera', 'Fagus Sylvatica', 'Ginkgo Biloba', 'Ilex Aquifolium', 'Ilex Cornuta', 'Liquidambar Styraciflua', 'Liriodendron Tulipifera', 'Lithocarpus Cleistocarpus', 'Lithocarpus Edulis', 'Magnolia Heptapeta', 'Magnolia Salicifolia', 'Morus Nigra', 'Olea Europaea', 'Phildelphus', 'Populus Adenopoda', 'Populus Grandidentata', 'Populus Nigra', 'Prunus Avium', 'Prunus X Shmittii', 'Pterocarya Stenoptera', 'Quercus Afares', 'Quercus Agrifolia', 'Quercus Alnifolia', 'Quercus Brantii', 'Quercus Canariensis', 'Quercus Castaneifolia', 'Quercus Cerris', 'Quercus Chrysolepis', 'Quercus Coccifera', 'Quercus Coccinea', 'Quercus Crassifolia', 'Quercus Crassipes', 'Quercus Dolicholepis', 'Quercus Ellipsoidalis', 'Quercus Greggii', 'Quercus Hartwissiana', 'Quercus Ilex', 'Quercus Imbricaria', 'Quercus Infectoria sub', 'Quercus Kewensis', 'Quercus Nigra', 'Quercus Palustris', 'Quercus Phellos', 'Quercus Phillyraeoides', 'Quercus Pontica', 'Quercus Pubescens', 'Quercus Pyrenaica', 'Quercus Rhysophylla', 'Quercus Rubra', 'Quercus Semecarpifolia', 'Quercus Shumardii', 'Quercus Suber', 'Quercus Texana', 'Quercus Trojana', 'Quercus Variabilis', 'Quercus Vulcanica', 'Quercus x Hispanica', 'Quercus x Turneri', 'Rhododendron x Russellianum', 'Salix Fragilis', 'Salix Intergra', 'Sorbus Aria', 'Tilia Oliveri', 'Tilia Platyphyllos', 'Tilia Tomentosa', 'Ulmus Bergmanniana', 'Viburnum Tinus', 'Viburnum x Rhytidophylloides', 'Zelkova Serrata'
##
## Pre-processing: Box-Cox transformation (64), centered (192), scaled
## (192), principal component signal extraction (192)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 1078, 1074, 1084, 1076, 1083, 1082, ...
## Resampling results:
##
## Accuracy Kappa
## 0.9832037 0.9830154
CopyTestset confusion matrix.
result$confusionMatrix$overall[1:4]
## Accuracy Kappa AccuracyLower AccuracyUpper
## 0.9900000 0.9898990 0.9745952 0.9972688
CopyWe take notice of accuracy for final report.
leaf_data_accuracy <- result$confusionMatrix$overall[1]
CopyFinal Results
Finally, we gather all results in a data frame and show for comparison. The V symbol indicates when a dataset is selected for model building.
LDA_accuracy = c(shape_data_accuracy, texture_data_accuracy, margin_data_accuracy,
shape_texture_data_accuracy, margin_shape_data_accuracy, margin_texture_data_accuracy,
leaf_data_accuracy)
LDA_accuracy <- 100*LDA_accuracy # as percentage
results <- data.frame(SHA = c("V", " ", " ", "V", "V", " ", "V"),
TEX = c(" ", "V", " ", "V", " ", "V", "V"),
MAR = c(" ", " ", "V", " ", "V", "V", "V"),
prop_KNN_accuracy = c(62.13, 72.94, 75.00, 86.19, 87.19, 93.38, 96.81),
wprop_KNN__accuracy = c(61.88, 72.75, 75.75, 86.06, 86.75, 93.31, 96.69),
LDA_accuracy = LDA_accuracy
)
CopyHere are ref.[1] results compared to ours.
results
## SHA TEX MAR prop_KNN_accuracy wprop_KNN__accuracy LDA_accuracy
## 1 V 62.13 61.88 51.50
## 2 V 72.94 72.75 75.75
## 3 V 75.00 75.75 79.50
## 4 V V 86.19 86.06 92.25
## 5 V V 87.19 86.75 93.00
## 6 V V 93.38 93.31 94.75
## 7 V V V 96.81 96.69 99.00
CopyAs we can see, with the only exception of the shape dataset scenario, for all other cases as defined by the datasets in use, Linear Discriminant Analysis achieves higher accuracy with respect ref. [1] K-Nearest-Neighbor based models.
References
- Charles Mallah, James Cope and James Orwell, “PLANT LEAF CLASSIFICATION USING PROBABILISTIC INTEGRATION OF SHAPE, TEXTURE AND MARGIN FEATURES” link
- Leaf Dataset
- Caret package – train models by tag
- Combining PCA and LDA
- Linear Discriminant Analysis
- Linear Discriminant Analysis – Bit by Bit