Missing data is a problem that challenges data analysis methodologically and computationally in medical research. Patients of the clinical trials and cohort studies may drop out of the study, and therefore, generate missing data. The missing data could be at random when participants who drop out of study are not different from those who remained in the study. For example, in the study of body mass index and cholesterol levels, participants who don’t measure their blood cholesterol have a comparable body mass index with participants who measure their blood cholesterol.
To handle missing data researchers often choose to conduct analysis among participants without missing data (i.e., complete case analysis), but sometimes they prefer to impute the data. In the previous tutorials (Tutorial 1, Tutorial 2) published at DataScience+ we have shown how to impute the missing data by using MICE package. In this tutorial I will show how to graphically present the missing data, with only one purpose, to find whether missing is at random. Therefore, we will build plots by using the function marginplot from VIM package. This would be a short “how to” tutorial and have no intention to explain types of the missing data. You can learn more about types of missing data such as missing completely at random, missing at random, and not missing at random from the book Statistical Analysis with Missing Data.
Data Preparation
I simulated a database with 250 observations for illustration purpose, and there is no clinical relevance. There are five variables Age, Gender, Cholesterol, SystolicBP, BMI, Smoking, and Education.
Load the libraries and get the data by running the scrip below:
library(VIM)
library(mice)
library(dplyr)
library(tibble)
dat <- read.csv(url("https://goo.gl/4DYzru"), header=TRUE, sep=",")
head(dat)
## Age Gender Cholesterol SystolicBP BMI Smoking Education
## 1 67.9 Female 236.4 129.8 26.4 Yes High
## 2 54.8 Female 256.3 133.4 28.4 No Medium
## 3 68.4 Male 198.7 158.5 24.1 Yes High
## 4 67.9 Male 205.0 136.0 19.9 No Low
## 5 60.9 Male 207.7 145.4 26.7 No Medium
## 6 44.9 Female 222.5 130.6 30.6 No LowIn this database, there are no missing. I will introduce missing not at random for the cholesterol variable. As is shown in the code below, missing values for cholesterol will include only participants with body mass index levels greater than 30 (i.e., participants with obesity).
set.seed(10) missing = rbinom(250, 1, 0.3) dat$Cholesterol = with(dat, ifelse(BMI>=30&missing==1, NA, Cholesterol)) sum(is.na(dat$Cholesterol)) [1] 16
We have 16 participants with missing values in cholesterol variable. I am going to impute the missing values by using MICE package and PMM (predictive mean matching) method.
init = mice(dat, maxit=0)
meth = init$method
predM = init$predictorMatrix
meth[c("Cholesterol")]="pmm"
set.seed(101)
imputed = mice(dat, method=meth, predictorMatrix=predM, m=1)
imp = complete(imputed)Next, I will create a database with imputed data and an indicator variable which shows which observations are imputed. This is necessary for plotting by using marginplot function.
dt1 = dat %>%
select(Cholesterol, BMI) %>%
rename(Cholesterol_imp = Cholesterol) %>%
mutate(
Cholesterol_imp = as.logical(ifelse(is.na(Cholesterol_imp), "TRUE", "FALSE"))
) %>%
rownames_to_column()
dt2 = imp %>%
select(Cholesterol, BMI) %>%
rownames_to_column()
dt = left_join(dt1, dt2)
head(dt)
rowname Cholesterol_imp BMI Cholesterol
1 1 FALSE 26.4 236.4
2 2 FALSE 28.4 256.3
3 3 FALSE 24.1 198.7
4 4 FALSE 19.9 205.0
5 5 FALSE 26.7 207.7
6 6 FALSE 30.6 222.5Graphical presentation of missing data
Now that we have a database with the imputed variables and the corresponding indicator, whether the observation is imputed or not, we will plot it by using the function marginplot from VIM package.
vars <- c("BMI","Cholesterol","Cholesterol_imp")
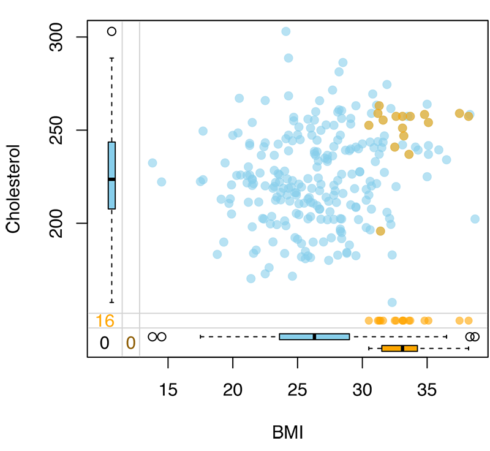
marginplot(dt[,vars], delimiter="_imp", alpha=0.6, pch=c(19))This is the output of code above:
The blue color in the scatterplot above indicate the non-missing values for the cholesterol, and the orange shows the missing data which is imputed. As we can see participants with missing data in cholesterol have a higher body mass index that participants without missing data which indicate that missing is not at random. This scatterplot also shows the boxplot which illustrates the distribution of data. For example, the median of body mass index for participants with missing data is around 33, whereas for those without missing is round 26.
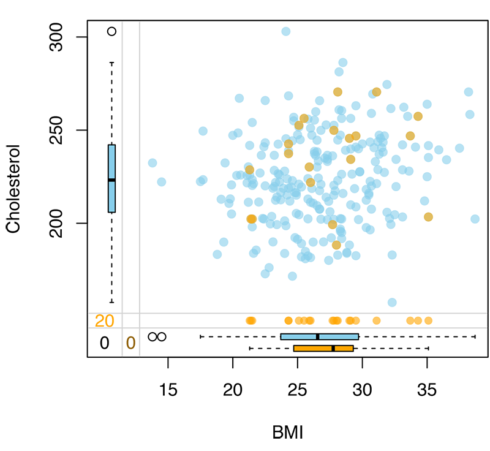
A missing at random will look like in the plot below
Conclusion
In this post, we showed how to use marginplot of VIM package to identify whether your missing data is at random. To learn more about VIM package I suggest to read the paper by Bernd Prantner.
I hope you find this post useful. Leave comments below if you have questions.